Night sky in Silver Peak Wilderness, California. Nov 24 '24
I am an applied machine learning researcher at NASA Ames, specialized in the application of machine learning to astronomy and astrophysics.
My research primarily focuses on enhancing the output of exoplanet survey missions, such as Kepler and TESS. I develop machine learning models to analyze high-quality, large datasets of transit signals, aiming to accelerate the vetting of planet candidates and the validation of exoplanets. Additionally, I have interests in uncertainty quantification, the characterization of deep learning models, and model explainability (XAI).
From more general topics...
Uncertainty quantification and characterization of deep learning models.
Model explainability (XAI).
...to more concrete applications:
Denoising mid-infrared images from the James Webb Space Telescope (JWST) by developing algorithms for cosmic ray detection, which facilitate the creation of high-quality data products from NASA's flagship observatory.
Performing transit detection for TESS and Kepler using machine learning methods.
Research
This is a list that intends to give an overview of my current and previous
research. For a comprehensive list of my published work you can see my
Google Scholar
page or find me on NASA ADS.
 ExoMiner Pipeline on NASA GitHub. Powered by Podman 🤖.
ExoMiner Pipeline on NASA GitHub. Powered by Podman 🤖.
End-to-end ExoMiner Pipeline for the TESS Mission 🛰️
ExoMiner on TESS has been integrated into an end-to-end pipeline that queries SPOC TCEs for
a set of TIC IDs, grabs and processes the light curve and image data from MAST and generates predictions
for the detected TCEs in those TIC IDs. Made with Podman to ease the use for any exoplanet hunter out there!
ExoMiner Pipeline on Podman
For those that want to tweak with the pipeline, the NASA GitHub repository provides access to the full codebase along with
the YAML file required to reproduce the Conda environment with all the 3rd party packages used in this project. "Watch" the repository
so you can be up-to-date on the latest advancements!
ExoMiner on NASA GitHub
 Artist's depiction of TESS. Credit: NASA/MIT.
Artist's depiction of TESS. Credit: NASA/MIT.
Vetting Transit Signals from the TESS Mission 🪐
We adapted ExoMiner to the TESS mission data to search through the
hundreds of thousands of transit signals generated by Primary, 1st,
and 2nd Extended Missions of TESS.
For each sector run and for both 2-min cadence and full-frame image data,
we generated catalogs of Threshold Crossing Events (TCEs) and Community
TESS Objects of Interest (CTOIs). These vetting catalogs
aim to provide the community with a more (limited) set of interesting
transit signals and make manual vetting more targeted and less
time-consuming.
ExoMiner on TESS 2-min [AJ]
ExoMiner Vetting Planet Candidate Catalog for S1-S67 TESS SPOC 2-min TCEs: Interactive Table
Other studies using TESS data include:
ExoMiner on TESS FFI Data [Ongoing Work - Soon!]| Presentation
 Artist's depiction of Kepler. Credit: NASA/ESA/CSA/STScI.
Artist's depiction of Kepler. Credit: NASA/ESA/CSA/STScI.
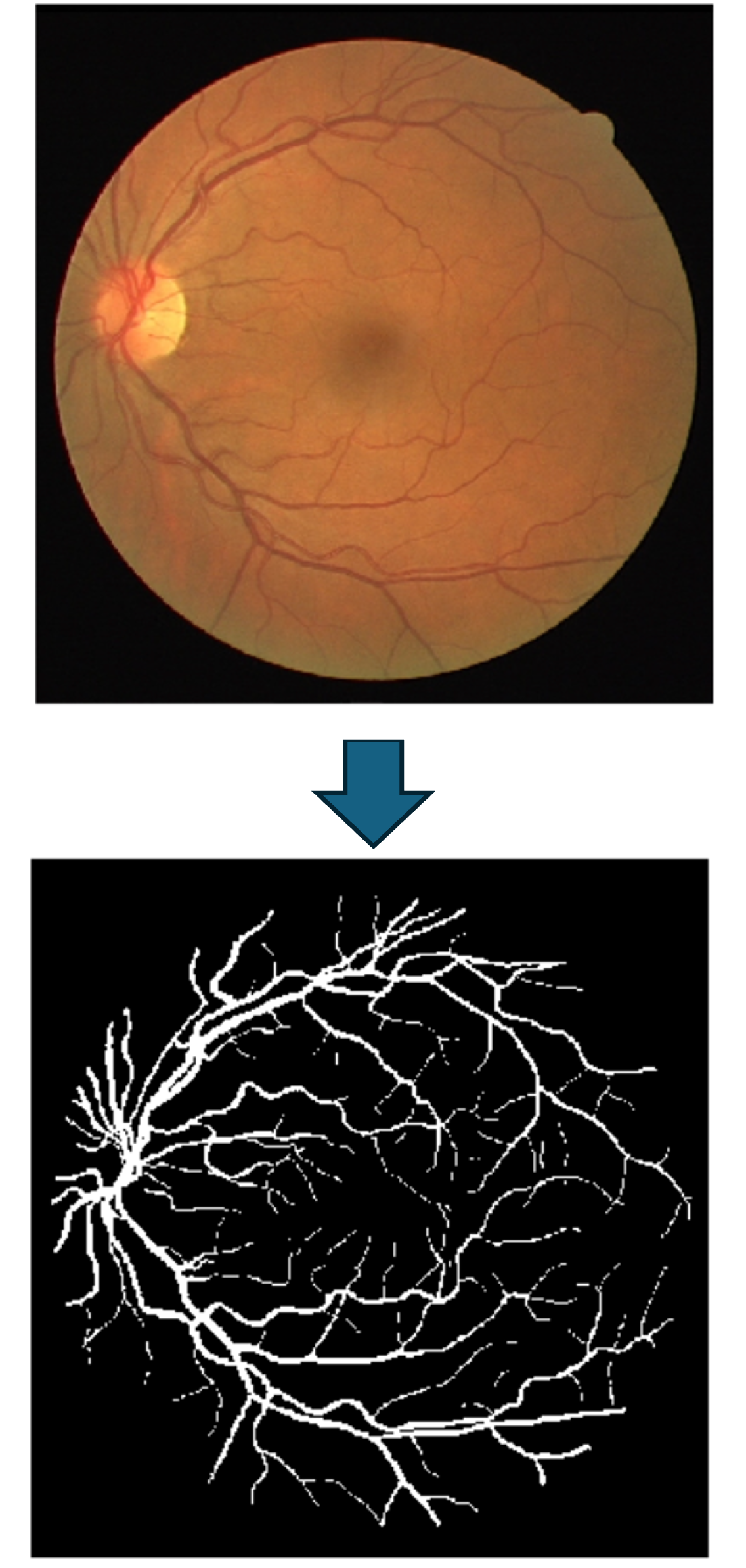 Credit: Example from the DRIVE dataset.
Credit: Example from the DRIVE dataset.
Vascular Tissue Segmentation for VESGEN 👁️
Developed convolutional-based models like U-Nets to perform the segmentation
of vascular networks and trees for VESGEN.
VESGEN is a NASA's software created to map vascular patterns in tissues
such as the human retina and quantify processes that include angiogenesis,
with the goal of enabling a better understanding of terrestrial
diseases as well as study the effects of microgravity and radiation
exposure to astronauts and other organisms in space environments.
Using a public dataset of human retina images (DRIVE ),
we extended our private dataset of vascular images of patients with different
degrees of diabetic retinopathy to train and evaluate the performance of our models.
An additional dataset of images from astronauts before and after
their missions to the ISS was also explored. This project started as
a proof of concept to perform the automatic binarization of vascular
patterning in the human retina to free researchers and clinicians
from the time-consuming task (3-20 hours) of producing a manual segmentation map.
Lagatuz et al. (2021)





